class: title-slide, left, top # Package Building ## with `devtools` and `usethis` ### Tom Mock & Josiah Parry ### 2021-10-21 <br> #### <svg aria-hidden="true" role="img" viewBox="0 0 512 512" style="height:1em;width:1em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:white;overflow:visible;position:relative;"><path d="M326.612 185.391c59.747 59.809 58.927 155.698.36 214.59-.11.12-.24.25-.36.37l-67.2 67.2c-59.27 59.27-155.699 59.262-214.96 0-59.27-59.26-59.27-155.7 0-214.96l37.106-37.106c9.84-9.84 26.786-3.3 27.294 10.606.648 17.722 3.826 35.527 9.69 52.721 1.986 5.822.567 12.262-3.783 16.612l-13.087 13.087c-28.026 28.026-28.905 73.66-1.155 101.96 28.024 28.579 74.086 28.749 102.325.51l67.2-67.19c28.191-28.191 28.073-73.757 0-101.83-3.701-3.694-7.429-6.564-10.341-8.569a16.037 16.037 0 0 1-6.947-12.606c-.396-10.567 3.348-21.456 11.698-29.806l21.054-21.055c5.521-5.521 14.182-6.199 20.584-1.731a152.482 152.482 0 0 1 20.522 17.197zM467.547 44.449c-59.261-59.262-155.69-59.27-214.96 0l-67.2 67.2c-.12.12-.25.25-.36.37-58.566 58.892-59.387 154.781.36 214.59a152.454 152.454 0 0 0 20.521 17.196c6.402 4.468 15.064 3.789 20.584-1.731l21.054-21.055c8.35-8.35 12.094-19.239 11.698-29.806a16.037 16.037 0 0 0-6.947-12.606c-2.912-2.005-6.64-4.875-10.341-8.569-28.073-28.073-28.191-73.639 0-101.83l67.2-67.19c28.239-28.239 74.3-28.069 102.325.51 27.75 28.3 26.872 73.934-1.155 101.96l-13.087 13.087c-4.35 4.35-5.769 10.79-3.783 16.612 5.864 17.194 9.042 34.999 9.69 52.721.509 13.906 17.454 20.446 27.294 10.606l37.106-37.106c59.271-59.259 59.271-155.699.001-214.959z"/></svg> [colorado.rstudio.com/rsc/pkg-building](https://colorado.rstudio.com/rsc/pkg-building/) #### <svg aria-hidden="true" role="img" viewBox="0 0 496 512" style="height:1em;width:0.97em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:white;overflow:visible;position:relative;"><path d="M165.9 397.4c0 2-2.3 3.6-5.2 3.6-3.3.3-5.6-1.3-5.6-3.6 0-2 2.3-3.6 5.2-3.6 3-.3 5.6 1.3 5.6 3.6zm-31.1-4.5c-.7 2 1.3 4.3 4.3 4.9 2.6 1 5.6 0 6.2-2s-1.3-4.3-4.3-5.2c-2.6-.7-5.5.3-6.2 2.3zm44.2-1.7c-2.9.7-4.9 2.6-4.6 4.9.3 2 2.9 3.3 5.9 2.6 2.9-.7 4.9-2.6 4.6-4.6-.3-1.9-3-3.2-5.9-2.9zM244.8 8C106.1 8 0 113.3 0 252c0 110.9 69.8 205.8 169.5 239.2 12.8 2.3 17.3-5.6 17.3-12.1 0-6.2-.3-40.4-.3-61.4 0 0-70 15-84.7-29.8 0 0-11.4-29.1-27.8-36.6 0 0-22.9-15.7 1.6-15.4 0 0 24.9 2 38.6 25.8 21.9 38.6 58.6 27.5 72.9 20.9 2.3-16 8.8-27.1 16-33.7-55.9-6.2-112.3-14.3-112.3-110.5 0-27.5 7.6-41.3 23.6-58.9-2.6-6.5-11.1-33.3 2.6-67.9 20.9-6.5 69 27 69 27 20-5.6 41.5-8.5 62.8-8.5s42.8 2.9 62.8 8.5c0 0 48.1-33.6 69-27 13.7 34.7 5.2 61.4 2.6 67.9 16 17.7 25.8 31.5 25.8 58.9 0 96.5-58.9 104.2-114.8 110.5 9.2 7.9 17 22.9 17 46.4 0 33.7-.3 75.4-.3 83.6 0 6.5 4.6 14.4 17.3 12.1C428.2 457.8 496 362.9 496 252 496 113.3 383.5 8 244.8 8zM97.2 352.9c-1.3 1-1 3.3.7 5.2 1.6 1.6 3.9 2.3 5.2 1 1.3-1 1-3.3-.7-5.2-1.6-1.6-3.9-2.3-5.2-1zm-10.8-8.1c-.7 1.3.3 2.9 2.3 3.9 1.6 1 3.6.7 4.3-.7.7-1.3-.3-2.9-2.3-3.9-2-.6-3.6-.3-4.3.7zm32.4 35.6c-1.6 1.3-1 4.3 1.3 6.2 2.3 2.3 5.2 2.6 6.5 1 1.3-1.3.7-4.3-1.3-6.2-2.2-2.3-5.2-2.6-6.5-1zm-11.4-14.7c-1.6 1-1.6 3.6 0 5.9 1.6 2.3 4.3 3.3 5.6 2.3 1.6-1.3 1.6-3.9 0-6.2-1.4-2.3-4-3.3-5.6-2z"/></svg> [github.com/jthomasmock/pkg-building](https://github.com/jthomasmock/pkg-building) <span style='color:white;'>Slides released under</span> [CC-BY 2.0](https://creativecommons.org/licenses/by/2.0/) <svg aria-hidden="true" role="img" viewBox="0 0 496 512" style="height:1em;width:0.97em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:white;overflow:visible;position:relative;"><path d="M245.83 214.87l-33.22 17.28c-9.43-19.58-25.24-19.93-27.46-19.93-22.13 0-33.22 14.61-33.22 43.84 0 23.57 9.21 43.84 33.22 43.84 14.47 0 24.65-7.09 30.57-21.26l30.55 15.5c-6.17 11.51-25.69 38.98-65.1 38.98-22.6 0-73.96-10.32-73.96-77.05 0-58.69 43-77.06 72.63-77.06 30.72-.01 52.7 11.95 65.99 35.86zm143.05 0l-32.78 17.28c-9.5-19.77-25.72-19.93-27.9-19.93-22.14 0-33.22 14.61-33.22 43.84 0 23.55 9.23 43.84 33.22 43.84 14.45 0 24.65-7.09 30.54-21.26l31 15.5c-2.1 3.75-21.39 38.98-65.09 38.98-22.69 0-73.96-9.87-73.96-77.05 0-58.67 42.97-77.06 72.63-77.06 30.71-.01 52.58 11.95 65.56 35.86zM247.56 8.05C104.74 8.05 0 123.11 0 256.05c0 138.49 113.6 248 247.56 248 129.93 0 248.44-100.87 248.44-248 0-137.87-106.62-248-248.44-248zm.87 450.81c-112.54 0-203.7-93.04-203.7-202.81 0-105.42 85.43-203.27 203.72-203.27 112.53 0 202.82 89.46 202.82 203.26-.01 121.69-99.68 202.82-202.84 202.82z"/></svg><svg aria-hidden="true" role="img" viewBox="0 0 496 512" style="height:1em;width:0.97em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:white;overflow:visible;position:relative;"><path d="M314.9 194.4v101.4h-28.3v120.5h-77.1V295.9h-28.3V194.4c0-4.4 1.6-8.2 4.6-11.3 3.1-3.1 6.9-4.7 11.3-4.7H299c4.1 0 7.8 1.6 11.1 4.7 3.1 3.2 4.8 6.9 4.8 11.3zm-101.5-63.7c0-23.3 11.5-35 34.5-35s34.5 11.7 34.5 35c0 23-11.5 34.5-34.5 34.5s-34.5-11.5-34.5-34.5zM247.6 8C389.4 8 496 118.1 496 256c0 147.1-118.5 248-248.4 248C113.6 504 0 394.5 0 256 0 123.1 104.7 8 247.6 8zm.8 44.7C130.2 52.7 44.7 150.6 44.7 256c0 109.8 91.2 202.8 203.7 202.8 103.2 0 202.8-81.1 202.8-202.8.1-113.8-90.2-203.3-202.8-203.3z"/></svg> <div style = "position: absolute;top: 0px;right: 0;"><img src="https://raw.githubusercontent.com/jthomasmock/pkg-building/master/images/pkg-love.png" alt="A person holding a Rubik's cube in their hand. The cube has a heart shaped in the middle." width="600"></img></div> --- layout: true <div class="my-footer"><span>https://colorado.rstudio.com/rsc/pkg-building/</span></div> --- ### Do you want to build a package? > Packages are the fundamental units of reproducible R code. They include reusable R functions, the documentation that describes how to use them, and sample data. > - [Hadley Wickham and Jenny Bryan](https://r-pkgs.org/index.html) -- In other words, a **package** is a home for *functions*. -- And **functions** are a home for *source code*. -- ### Stating this a bit differently: -- Functions in R are _just_ wrappers around longer source code. -- Packages are _just_ a way of **describing** and **distributing** these functions, in a **structured and consistent way**. --- ### Do you want to build a ~~package~~ home for functions? > Reproducibility is actually all about being as lazy as possible! > – Hadley Wickham -- ### Functions are a way to: -- * Don't repeat yourself (DRY principle) and be more efficient * Share workflows and empower yourself and your team * Test your code and "trust your work", or trust _others_ work -- Ultimately, functions make your work much easier, faster, and more reproducible -- ...and R packages let you share these functions and be lazier, in a good way! --- class: inverse, center, middle # Functions --- ### Anatomy of a Function 1. A descriptive function **name**, informing the user of the function's purpose -- 2. The argument(s) to the **function**, controlling the output of the function -- 3. The **body** of the function, that represents all the code to be used internally -- 4. What the function **returns** -- ```r my_function <- function(argument){ # fancy_code goes here # using the **argument** # to do something cool # **returns** a result } ``` --- ### Our first `function` A simple function, with: * one **argument** `x` * that takes `x` and squares it * and then **returns** it -- ```r square_val <- function(x){ x^2 } ``` -- ```r square_val(2) ``` ``` ## [1] 4 ``` ```r square_val(16) ``` ``` ## [1] 256 ``` --- ### Our first `function` A simple function, with: * one **argument** `x` * that takes `x` and squares it * and then **returns** it But what about unintended inputs? -- ```r "cat"^2 ``` ``` ## Error in "cat"^2: non-numeric argument to binary operator ``` ```r square_val("cat") ``` ``` ## Error in x^2: non-numeric argument to binary operator ``` --- ### Our first `function2` ```r square_val2 <- function(x){ stopifnot("Input must be numeric" = is.numeric(x)) x^2 } ``` -- ```r square_val2("cat") ``` ``` ## Error in square_val2("cat"): Input must be numeric ``` ```r square_val2(3) ``` ``` ## [1] 9 ``` --- ### Generate fake data, reproducibly ```r generate_df <- function(n = 10, with_seed = NULL){ # If a seed is specified, then use it, otherwise ignore if(!is.null(with_seed)){set.seed(with_seed)} # pad the values with repeated zeros pad_length <- paste0("%0", nchar(n), "d") random_int <- sample(1:n, replace = TRUE) padded_int <- sprintf(pad_length, random_int) # create a df with combined random letters and integers tibble::tibble( id = paste0( sample(LETTERS, size = n, replace = TRUE), padded_int ), values = rnorm(n = n, mean = 15, sd = 2) ) } ``` --- ### Generate fake data, reproducibly ```r generate_df(125) ``` ``` ## # A tibble: 125 × 2 ## id values ## <chr> <dbl> ## 1 Q096 16.0 ## 2 W007 13.1 ## 3 R030 13.7 ## 4 Q013 17.9 ## 5 K073 13.4 ## 6 S086 12.3 ## 7 I018 11.0 ## 8 L064 16.2 ## 9 F025 14.9 ## 10 U024 18.8 ## # … with 115 more rows ``` --- ### Reproducibly random ```r all.equal( generate_df(10, with_seed = 123), generate_df(10, with_seed = 123) ) ``` ``` ## [1] TRUE ``` -- ### Or fully random ```r all.equal( generate_df(10), generate_df(10) ) ``` ``` ## [1] "Component \"id\": 10 string mismatches" ## [2] "Component \"values\": Mean relative difference: 0.1569146" ``` --- ### Fake data, with customization ```r generate_df2 <- function(n = 10, with_seed = NULL, ...){ # If a seed is specified, then use it, otherwise ignore if(!is.null(with_seed)){set.seed(with_seed)} # pad the values with repeated zeros pad_length <- paste0("%0", nchar(n), "d") random_int <- sample(1:n, replace = TRUE) padded_int <- sprintf(pad_length, random_int) # create a df with combined random letters and integers tibble::tibble( id = paste0(sample(LETTERS, n, replace = TRUE), padded_int), values = rnorm(n = n, ...) ) } ``` --- ### Fake data, with customization ```r generate_df2(125, with_seed = 37, mean = 10, sd = 2) ``` ``` ## # A tibble: 125 × 2 ## id values ## <chr> <dbl> ## 1 L054 10.2 ## 2 Y047 11.2 ## 3 Q024 12.7 ## 4 V050 9.61 ## 5 V003 11.1 ## 6 X104 8.84 ## 7 H109 7.73 ## 8 G036 9.49 ## 9 E041 11.3 ## 10 Z052 12.0 ## # … with 115 more rows ``` --- ### Actual use in `gtExtras` ```r generate_df <- function(n = 10L, n_grps = 1L, mean = c(10), sd = mean/10, with_seed = NULL){ # If a seed is specified, then use it, otherwise ignore if(!base::is.null(with_seed)){base::set.seed(with_seed)} # pad the values with repeated zeros pad_length <- base::paste0("%0", nchar(n), "d") random_int <- base::sample(1:n, replace = TRUE) padded_int <- base::sprintf(pad_length, random_int) # create a df with combined random letters and integers dplyr::tibble( row_id = 1:(n*n_grps), id = paste0(sample(LETTERS, n*n_grps, replace = TRUE), padded_int), grp = sprintf("grp-%s", 1:n_grps) %>% rep(each = n), values = mapply(rnorm, n, mean, sd) %>% as.vector() ) } ``` --- ### Actual use in `gtExtras` ```r generate_df() ``` ``` ## # A tibble: 10 × 4 ## row_id id grp values ## <int> <chr> <chr> <dbl> ## 1 1 E05 grp-1 10.5 ## 2 2 A09 grp-1 9.73 ## 3 3 C07 grp-1 10.5 ## 4 4 L01 grp-1 10.9 ## 5 5 A08 grp-1 8.80 ## 6 6 C01 grp-1 10.1 ## 7 7 Z07 grp-1 9.48 ## 8 8 E08 grp-1 10.0 ## 9 9 T01 grp-1 9.77 ## 10 10 L07 grp-1 10.9 ``` --- ### Don't repeat yourself (Sure, you could use `regex` or even `stringr`, but let's not for now) ```r my_string <- "RStudio 147" substr(my_string, 9, 11) ``` ``` ## [1] "147" ``` -- ```r substr("Wales National 148", 9, 11) ``` ``` ## [1] "tio" ``` -- ```r substr("Wales National 148", -3, -1) ``` ``` ## [1] "" ``` --- ```r substr_right <- function(x, n){ char_count <- nchar(x) sub_n <- n - 1 substr(x, char_count-sub_n, char_count) } ``` -- ```r substr_right("Wales National 148", 3) ``` ``` ## [1] "148" ``` -- ```r substr_right("RStudio 147", 3) ``` ``` ## [1] "147" ``` --- ```r set.seed(73) company_df <- tibble::tibble( x = 1:10, company_id = paste( c("RStudio", "Wales National", "Widget Corp", "Product Co", "Food, Inc", "DairyCo", "TechMart", "Gas Giant", "Consulting United", "Math Magic"), sample(100:999, size = 10) ), sales = rnorm(10, mean = 15000, sd = 1000) ) company_df ``` ``` ## # A tibble: 10 × 3 ## x company_id sales ## <int> <chr> <dbl> ## 1 1 RStudio 416 15628. ## 2 2 Wales National 426 15713. ## 3 3 Widget Corp 443 16373. ## 4 4 Product Co 980 16428. ## 5 5 Food, Inc 500 12753. ## 6 6 DairyCo 914 14076. ## 7 7 TechMart 874 13862. ## 8 8 Gas Giant 374 14887. ## 9 9 Consulting United 823 16286. ## 10 10 Math Magic 706 15410. ``` --- ```r company_df %>% mutate(id_num = substr_right(company_id, 3)) ``` -- ``` ## # A tibble: 10 × 4 ## x company_id sales id_num ## <int> <chr> <dbl> <chr> ## 1 1 RStudio 416 15628. 416 ## 2 2 Wales National 426 15713. 426 ## 3 3 Widget Corp 443 16373. 443 ## 4 4 Product Co 980 16428. 980 ## 5 5 Food, Inc 500 12753. 500 ## 6 6 DairyCo 914 14076. 914 ## 7 7 TechMart 874 13862. 874 ## 8 8 Gas Giant 374 14887. 374 ## 9 9 Consulting United 823 16286. 823 ## 10 10 Math Magic 706 15410. 706 ``` --- class: center, middle, inverse # `tidyeval` functions --- ### A short primer on `tidyeval` > Most dplyr verbs use **tidy evaluation** in some way. Tidy evaluation is a special type of non-standard evaluation used throughout the tidyverse. - [Programming with `dplyr`](https://dplyr.tidyverse.org/articles/programming.html) -- This powers the ability to do: ``` mtcars %>% # Knows where and how to find `cyl` as a column, not an object group_by(cyl) %>% # knows how to find `mpg` as a column, not an object summarize(n = n(), mean = mean(mpg)) ``` -- In my opinion, you can get most `tidyeval` things working with just two new concepts: * Embrace your variable with `{{ var }}`, also known as 'curly-curly' * Pass the dots with `...` for many arguments -- And you can always revert back to `.data[[var]]` if you want to use `"strings"` instead of bare `columns` --- ### `tidyeval` in practice .pull-left[ ```r library(dplyr) car_summary <- function(var){ mtcars %>% group_by({{var}}) %>% summarize(mean = mean(mpg), n = n()) } ``` ] -- .pull-right[ ```r car_summary(vs) ``` ``` ## # A tibble: 2 × 3 ## vs mean n ## <dbl> <dbl> <int> ## 1 0 16.6 18 ## 2 1 24.6 14 ``` ] -- ```r car_summary(vs, am) ``` ``` ## Error in car_summary(vs, am): unused argument (am) ``` --- ### `tidyeval` in practice ```r library(dplyr) car_summary_dots <- function(...){ mtcars %>% # add the dots group_by(...) %>% summarize(mean = mean(mpg), n = n(), .groups = "drop") } ``` -- ```r car_summary_dots(vs, am, cyl) ``` ``` ## # A tibble: 7 × 5 ## vs am cyl mean n ## <dbl> <dbl> <dbl> <dbl> <int> ## 1 0 0 8 15.0 12 ## 2 0 1 4 26 1 ## 3 0 1 6 20.6 3 ## 4 0 1 8 15.4 2 ## 5 1 0 4 22.9 3 ## 6 1 0 6 19.1 4 ## 7 1 1 4 28.4 7 ``` --- ### `tidyeval` in practice with optional arguments ```r library(dplyr) my_car_summary_dots2 <- function(var, ...){ mtcars %>% group_by({{var}}) %>% summarize(mean = mean(mpg), n = n(), ..., .groups = "drop") } ``` -- ```r my_car_summary_dots2(cyl, hp_mean = mean(hp), hp_sd = sd(hp)) ``` ``` ## # A tibble: 3 × 5 ## cyl mean n hp_mean hp_sd ## <dbl> <dbl> <int> <dbl> <dbl> ## 1 4 26.7 11 82.6 20.9 ## 2 6 19.7 7 122. 24.3 ## 3 8 15.1 14 209. 51.0 ``` --- ### `tidyeval` with novel data .pull-left[ ```r var_summary <- function(data, var) { data %>% summarise( n = n(), min = min({{ var }}), max = max({{ var }}), .groups = "drop") } ``` ```r mtcars %>% group_by(cyl) %>% var_summary(mpg) ``` ``` ## # A tibble: 3 × 4 ## cyl n min max ## <dbl> <int> <dbl> <dbl> ## 1 4 11 21.4 33.9 ## 2 6 7 17.8 21.4 ## 3 8 14 10.4 19.2 ``` ] -- .pull-right[ ```r ToothGrowth %>% group_by(supp, dose) %>% var_summary(len) ``` ``` ## # A tibble: 6 × 5 ## supp dose n min max ## <fct> <dbl> <int> <dbl> <dbl> ## 1 OJ 0.5 10 8.2 21.5 ## 2 OJ 1 10 14.5 27.3 ## 3 OJ 2 10 22.4 30.9 ## 4 VC 0.5 10 4.2 11.5 ## 5 VC 1 10 13.6 22.5 ## 6 VC 2 10 18.5 33.9 ``` ] --- ### `tidyeval` with `.data` and `"strings"` ```r var_summary2 <- function(data, var) { data %>% summarise(n = n(), min = min(.data[[var]]), max = max(.data[[var]]), .groups = "drop") } ``` -- ```r mtcars %>% group_by(cyl) %>% var_summary2("mpg") ``` ``` ## # A tibble: 3 × 4 ## cyl n min max ## <dbl> <int> <dbl> <dbl> ## 1 4 11 21.4 33.9 ## 2 6 7 17.8 21.4 ## 3 8 14 10.4 19.2 ``` --- ### One more `tidyeval` with `gt` * [`gtExtras::gt_add_divider()`](https://jthomasmock.github.io/gtExtras/reference/gt_add_divider.html), simplified ```r library(gt) gt_add_divider <- function(gt_object, columns, ..., include_labels = TRUE) { stopifnot("Table must be of class 'gt_tbl'" = "gt_tbl" %in% class(gt_object)) gt_object %>% tab_style( # dots include passed named arguments to the internal function style = cell_borders(sides = "right", ...), locations = if (isTRUE(include_labels)) { # columns to affect list(cells_body(columns = {{ columns }}), cells_column_labels(columns = {{ columns }})) } else { cells_body(columns = {{ columns }}) } ) } ``` --- ### One more `tidyeval` with `gt` ```r basic_table <- head(mtcars, 6) %>% gt() basic_table ``` <div id="tjiwclykiq" style="overflow-x:auto;overflow-y:auto;width:auto;height:auto;"> <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #tjiwclykiq .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #tjiwclykiq .gt_heading { background-color: #FFFFFF; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #tjiwclykiq .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #tjiwclykiq .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 6px; border-top-color: #FFFFFF; border-top-width: 0; } #tjiwclykiq .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #tjiwclykiq .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #tjiwclykiq .gt_col_heading { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #tjiwclykiq .gt_column_spanner_outer { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #tjiwclykiq .gt_column_spanner_outer:first-child { padding-left: 0; } #tjiwclykiq .gt_column_spanner_outer:last-child { padding-right: 0; } #tjiwclykiq .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 5px; overflow-x: hidden; display: inline-block; width: 100%; } #tjiwclykiq .gt_group_heading { padding: 8px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; } #tjiwclykiq .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #tjiwclykiq .gt_from_md > :first-child { margin-top: 0; } #tjiwclykiq .gt_from_md > :last-child { margin-bottom: 0; } #tjiwclykiq .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #tjiwclykiq .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 12px; } #tjiwclykiq .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #tjiwclykiq .gt_first_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; } #tjiwclykiq .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #tjiwclykiq .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #tjiwclykiq .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #tjiwclykiq .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #tjiwclykiq .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #tjiwclykiq .gt_footnote { margin: 0px; font-size: 90%; padding: 4px; } #tjiwclykiq .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #tjiwclykiq .gt_sourcenote { font-size: 90%; padding: 4px; } #tjiwclykiq .gt_left { text-align: left; } #tjiwclykiq .gt_center { text-align: center; } #tjiwclykiq .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #tjiwclykiq .gt_font_normal { font-weight: normal; } #tjiwclykiq .gt_font_bold { font-weight: bold; } #tjiwclykiq .gt_font_italic { font-style: italic; } #tjiwclykiq .gt_super { font-size: 65%; } #tjiwclykiq .gt_footnote_marks { font-style: italic; font-weight: normal; font-size: 65%; } </style> <table class="gt_table"> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">mpg</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">cyl</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">disp</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">hp</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">drat</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">wt</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">qsec</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">vs</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">am</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">gear</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">carb</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td class="gt_row gt_right">21.0</td> <td class="gt_row gt_right">6</td> <td class="gt_row gt_right">160</td> <td class="gt_row gt_right">110</td> <td class="gt_row gt_right">3.90</td> <td class="gt_row gt_right">2.620</td> <td class="gt_row gt_right">16.46</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">4</td> <td class="gt_row gt_right">4</td></tr> <tr><td class="gt_row gt_right">21.0</td> <td class="gt_row gt_right">6</td> <td class="gt_row gt_right">160</td> <td class="gt_row gt_right">110</td> <td class="gt_row gt_right">3.90</td> <td class="gt_row gt_right">2.875</td> <td class="gt_row gt_right">17.02</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">4</td> <td class="gt_row gt_right">4</td></tr> <tr><td class="gt_row gt_right">22.8</td> <td class="gt_row gt_right">4</td> <td class="gt_row gt_right">108</td> <td class="gt_row gt_right">93</td> <td class="gt_row gt_right">3.85</td> <td class="gt_row gt_right">2.320</td> <td class="gt_row gt_right">18.61</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">4</td> <td class="gt_row gt_right">1</td></tr> <tr><td class="gt_row gt_right">21.4</td> <td class="gt_row gt_right">6</td> <td class="gt_row gt_right">258</td> <td class="gt_row gt_right">110</td> <td class="gt_row gt_right">3.08</td> <td class="gt_row gt_right">3.215</td> <td class="gt_row gt_right">19.44</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">3</td> <td class="gt_row gt_right">1</td></tr> <tr><td class="gt_row gt_right">18.7</td> <td class="gt_row gt_right">8</td> <td class="gt_row gt_right">360</td> <td class="gt_row gt_right">175</td> <td class="gt_row gt_right">3.15</td> <td class="gt_row gt_right">3.440</td> <td class="gt_row gt_right">17.02</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">3</td> <td class="gt_row gt_right">2</td></tr> <tr><td class="gt_row gt_right">18.1</td> <td class="gt_row gt_right">6</td> <td class="gt_row gt_right">225</td> <td class="gt_row gt_right">105</td> <td class="gt_row gt_right">2.76</td> <td class="gt_row gt_right">3.460</td> <td class="gt_row gt_right">20.22</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">3</td> <td class="gt_row gt_right">1</td></tr> </tbody> </table> </div> --- ### One more `tidyeval` with `gt` ```r basic_table %>% # %>% data passed to 1st argument gt_add_divider(cyl) ``` <div id="fuwqugqdzp" style="overflow-x:auto;overflow-y:auto;width:auto;height:auto;"> <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #fuwqugqdzp .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #fuwqugqdzp .gt_heading { background-color: #FFFFFF; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #fuwqugqdzp .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #fuwqugqdzp .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 6px; border-top-color: #FFFFFF; border-top-width: 0; } #fuwqugqdzp .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #fuwqugqdzp .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #fuwqugqdzp .gt_col_heading { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #fuwqugqdzp .gt_column_spanner_outer { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #fuwqugqdzp .gt_column_spanner_outer:first-child { padding-left: 0; } #fuwqugqdzp .gt_column_spanner_outer:last-child { padding-right: 0; } #fuwqugqdzp .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 5px; overflow-x: hidden; display: inline-block; width: 100%; } #fuwqugqdzp .gt_group_heading { padding: 8px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; } #fuwqugqdzp .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #fuwqugqdzp .gt_from_md > :first-child { margin-top: 0; } #fuwqugqdzp .gt_from_md > :last-child { margin-bottom: 0; } #fuwqugqdzp .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #fuwqugqdzp .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 12px; } #fuwqugqdzp .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #fuwqugqdzp .gt_first_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; } #fuwqugqdzp .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #fuwqugqdzp .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #fuwqugqdzp .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #fuwqugqdzp .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #fuwqugqdzp .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #fuwqugqdzp .gt_footnote { margin: 0px; font-size: 90%; padding: 4px; } #fuwqugqdzp .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #fuwqugqdzp .gt_sourcenote { font-size: 90%; padding: 4px; } #fuwqugqdzp .gt_left { text-align: left; } #fuwqugqdzp .gt_center { text-align: center; } #fuwqugqdzp .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #fuwqugqdzp .gt_font_normal { font-weight: normal; } #fuwqugqdzp .gt_font_bold { font-weight: bold; } #fuwqugqdzp .gt_font_italic { font-style: italic; } #fuwqugqdzp .gt_super { font-size: 65%; } #fuwqugqdzp .gt_footnote_marks { font-style: italic; font-weight: normal; font-size: 65%; } </style> <table class="gt_table"> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">mpg</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" style="border-right-width: 1px; border-right-style: solid; border-right-color: #000000;">cyl</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">disp</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">hp</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">drat</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">wt</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">qsec</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">vs</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">am</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">gear</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">carb</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td class="gt_row gt_right">21.0</td> <td class="gt_row gt_right" style="border-right-width: 1px; border-right-style: solid; border-right-color: #000000;">6</td> <td class="gt_row gt_right">160</td> <td class="gt_row gt_right">110</td> <td class="gt_row gt_right">3.90</td> <td class="gt_row gt_right">2.620</td> <td class="gt_row gt_right">16.46</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">4</td> <td class="gt_row gt_right">4</td></tr> <tr><td class="gt_row gt_right">21.0</td> <td class="gt_row gt_right" style="border-right-width: 1px; border-right-style: solid; border-right-color: #000000;">6</td> <td class="gt_row gt_right">160</td> <td class="gt_row gt_right">110</td> <td class="gt_row gt_right">3.90</td> <td class="gt_row gt_right">2.875</td> <td class="gt_row gt_right">17.02</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">4</td> <td class="gt_row gt_right">4</td></tr> <tr><td class="gt_row gt_right">22.8</td> <td class="gt_row gt_right" style="border-right-width: 1px; border-right-style: solid; border-right-color: #000000;">4</td> <td class="gt_row gt_right">108</td> <td class="gt_row gt_right">93</td> <td class="gt_row gt_right">3.85</td> <td class="gt_row gt_right">2.320</td> <td class="gt_row gt_right">18.61</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">4</td> <td class="gt_row gt_right">1</td></tr> <tr><td class="gt_row gt_right">21.4</td> <td class="gt_row gt_right" style="border-right-width: 1px; border-right-style: solid; border-right-color: #000000;">6</td> <td class="gt_row gt_right">258</td> <td class="gt_row gt_right">110</td> <td class="gt_row gt_right">3.08</td> <td class="gt_row gt_right">3.215</td> <td class="gt_row gt_right">19.44</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">3</td> <td class="gt_row gt_right">1</td></tr> <tr><td class="gt_row gt_right">18.7</td> <td class="gt_row gt_right" style="border-right-width: 1px; border-right-style: solid; border-right-color: #000000;">8</td> <td class="gt_row gt_right">360</td> <td class="gt_row gt_right">175</td> <td class="gt_row gt_right">3.15</td> <td class="gt_row gt_right">3.440</td> <td class="gt_row gt_right">17.02</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">3</td> <td class="gt_row gt_right">2</td></tr> <tr><td class="gt_row gt_right">18.1</td> <td class="gt_row gt_right" style="border-right-width: 1px; border-right-style: solid; border-right-color: #000000;">6</td> <td class="gt_row gt_right">225</td> <td class="gt_row gt_right">105</td> <td class="gt_row gt_right">2.76</td> <td class="gt_row gt_right">3.460</td> <td class="gt_row gt_right">20.22</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">3</td> <td class="gt_row gt_right">1</td></tr> </tbody> </table> </div> --- ### One more `tidyeval` with `gt` ```r basic_table %>% # optional arguments accepted by name via `...` gt_add_divider(cyl, weight = px(2), color = "red") ``` <div id="oztqiatahj" style="overflow-x:auto;overflow-y:auto;width:auto;height:auto;"> <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #oztqiatahj .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #oztqiatahj .gt_heading { background-color: #FFFFFF; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #oztqiatahj .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #oztqiatahj .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 6px; border-top-color: #FFFFFF; border-top-width: 0; } #oztqiatahj .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #oztqiatahj .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #oztqiatahj .gt_col_heading { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #oztqiatahj .gt_column_spanner_outer { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #oztqiatahj .gt_column_spanner_outer:first-child { padding-left: 0; } #oztqiatahj .gt_column_spanner_outer:last-child { padding-right: 0; } #oztqiatahj .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 5px; overflow-x: hidden; display: inline-block; width: 100%; } #oztqiatahj .gt_group_heading { padding: 8px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; } #oztqiatahj .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #oztqiatahj .gt_from_md > :first-child { margin-top: 0; } #oztqiatahj .gt_from_md > :last-child { margin-bottom: 0; } #oztqiatahj .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #oztqiatahj .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 12px; } #oztqiatahj .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #oztqiatahj .gt_first_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; } #oztqiatahj .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #oztqiatahj .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #oztqiatahj .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #oztqiatahj .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #oztqiatahj .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #oztqiatahj .gt_footnote { margin: 0px; font-size: 90%; padding: 4px; } #oztqiatahj .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #oztqiatahj .gt_sourcenote { font-size: 90%; padding: 4px; } #oztqiatahj .gt_left { text-align: left; } #oztqiatahj .gt_center { text-align: center; } #oztqiatahj .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #oztqiatahj .gt_font_normal { font-weight: normal; } #oztqiatahj .gt_font_bold { font-weight: bold; } #oztqiatahj .gt_font_italic { font-style: italic; } #oztqiatahj .gt_super { font-size: 65%; } #oztqiatahj .gt_footnote_marks { font-style: italic; font-weight: normal; font-size: 65%; } </style> <table class="gt_table"> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">mpg</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1" style="border-right-width: 2px; border-right-style: solid; border-right-color: red;">cyl</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">disp</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">hp</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">drat</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">wt</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">qsec</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">vs</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">am</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">gear</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">carb</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td class="gt_row gt_right">21.0</td> <td class="gt_row gt_right" style="border-right-width: 2px; border-right-style: solid; border-right-color: red;">6</td> <td class="gt_row gt_right">160</td> <td class="gt_row gt_right">110</td> <td class="gt_row gt_right">3.90</td> <td class="gt_row gt_right">2.620</td> <td class="gt_row gt_right">16.46</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">4</td> <td class="gt_row gt_right">4</td></tr> <tr><td class="gt_row gt_right">21.0</td> <td class="gt_row gt_right" style="border-right-width: 2px; border-right-style: solid; border-right-color: red;">6</td> <td class="gt_row gt_right">160</td> <td class="gt_row gt_right">110</td> <td class="gt_row gt_right">3.90</td> <td class="gt_row gt_right">2.875</td> <td class="gt_row gt_right">17.02</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">4</td> <td class="gt_row gt_right">4</td></tr> <tr><td class="gt_row gt_right">22.8</td> <td class="gt_row gt_right" style="border-right-width: 2px; border-right-style: solid; border-right-color: red;">4</td> <td class="gt_row gt_right">108</td> <td class="gt_row gt_right">93</td> <td class="gt_row gt_right">3.85</td> <td class="gt_row gt_right">2.320</td> <td class="gt_row gt_right">18.61</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">4</td> <td class="gt_row gt_right">1</td></tr> <tr><td class="gt_row gt_right">21.4</td> <td class="gt_row gt_right" style="border-right-width: 2px; border-right-style: solid; border-right-color: red;">6</td> <td class="gt_row gt_right">258</td> <td class="gt_row gt_right">110</td> <td class="gt_row gt_right">3.08</td> <td class="gt_row gt_right">3.215</td> <td class="gt_row gt_right">19.44</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">3</td> <td class="gt_row gt_right">1</td></tr> <tr><td class="gt_row gt_right">18.7</td> <td class="gt_row gt_right" style="border-right-width: 2px; border-right-style: solid; border-right-color: red;">8</td> <td class="gt_row gt_right">360</td> <td class="gt_row gt_right">175</td> <td class="gt_row gt_right">3.15</td> <td class="gt_row gt_right">3.440</td> <td class="gt_row gt_right">17.02</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">3</td> <td class="gt_row gt_right">2</td></tr> <tr><td class="gt_row gt_right">18.1</td> <td class="gt_row gt_right" style="border-right-width: 2px; border-right-style: solid; border-right-color: red;">6</td> <td class="gt_row gt_right">225</td> <td class="gt_row gt_right">105</td> <td class="gt_row gt_right">2.76</td> <td class="gt_row gt_right">3.460</td> <td class="gt_row gt_right">20.22</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">3</td> <td class="gt_row gt_right">1</td></tr> </tbody> </table> </div> --- ### One more `tidyeval` with `gt` ```r basic_table %>% ### include_labels as an existing argument gt_add_divider(c(cyl,mpg), weight = px(3), color = "lightgrey", include_labels = FALSE) ``` <div id="gklsbnjpsx" style="overflow-x:auto;overflow-y:auto;width:auto;height:auto;"> <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #gklsbnjpsx .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #gklsbnjpsx .gt_heading { background-color: #FFFFFF; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #gklsbnjpsx .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #gklsbnjpsx .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 6px; border-top-color: #FFFFFF; border-top-width: 0; } #gklsbnjpsx .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #gklsbnjpsx .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #gklsbnjpsx .gt_col_heading { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #gklsbnjpsx .gt_column_spanner_outer { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #gklsbnjpsx .gt_column_spanner_outer:first-child { padding-left: 0; } #gklsbnjpsx .gt_column_spanner_outer:last-child { padding-right: 0; } #gklsbnjpsx .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 5px; overflow-x: hidden; display: inline-block; width: 100%; } #gklsbnjpsx .gt_group_heading { padding: 8px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; } #gklsbnjpsx .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #gklsbnjpsx .gt_from_md > :first-child { margin-top: 0; } #gklsbnjpsx .gt_from_md > :last-child { margin-bottom: 0; } #gklsbnjpsx .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #gklsbnjpsx .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 12px; } #gklsbnjpsx .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #gklsbnjpsx .gt_first_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; } #gklsbnjpsx .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #gklsbnjpsx .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #gklsbnjpsx .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #gklsbnjpsx .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #gklsbnjpsx .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #gklsbnjpsx .gt_footnote { margin: 0px; font-size: 90%; padding: 4px; } #gklsbnjpsx .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #gklsbnjpsx .gt_sourcenote { font-size: 90%; padding: 4px; } #gklsbnjpsx .gt_left { text-align: left; } #gklsbnjpsx .gt_center { text-align: center; } #gklsbnjpsx .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #gklsbnjpsx .gt_font_normal { font-weight: normal; } #gklsbnjpsx .gt_font_bold { font-weight: bold; } #gklsbnjpsx .gt_font_italic { font-style: italic; } #gklsbnjpsx .gt_super { font-size: 65%; } #gklsbnjpsx .gt_footnote_marks { font-style: italic; font-weight: normal; font-size: 65%; } </style> <table class="gt_table"> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">mpg</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">cyl</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">disp</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">hp</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">drat</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">wt</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">qsec</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">vs</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">am</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">gear</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">carb</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td class="gt_row gt_right" style="border-right-width: 3px; border-right-style: solid; border-right-color: lightgrey;">21.0</td> <td class="gt_row gt_right" style="border-right-width: 3px; border-right-style: solid; border-right-color: lightgrey;">6</td> <td class="gt_row gt_right">160</td> <td class="gt_row gt_right">110</td> <td class="gt_row gt_right">3.90</td> <td class="gt_row gt_right">2.620</td> <td class="gt_row gt_right">16.46</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">4</td> <td class="gt_row gt_right">4</td></tr> <tr><td class="gt_row gt_right" style="border-right-width: 3px; border-right-style: solid; border-right-color: lightgrey;">21.0</td> <td class="gt_row gt_right" style="border-right-width: 3px; border-right-style: solid; border-right-color: lightgrey;">6</td> <td class="gt_row gt_right">160</td> <td class="gt_row gt_right">110</td> <td class="gt_row gt_right">3.90</td> <td class="gt_row gt_right">2.875</td> <td class="gt_row gt_right">17.02</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">4</td> <td class="gt_row gt_right">4</td></tr> <tr><td class="gt_row gt_right" style="border-right-width: 3px; border-right-style: solid; border-right-color: lightgrey;">22.8</td> <td class="gt_row gt_right" style="border-right-width: 3px; border-right-style: solid; border-right-color: lightgrey;">4</td> <td class="gt_row gt_right">108</td> <td class="gt_row gt_right">93</td> <td class="gt_row gt_right">3.85</td> <td class="gt_row gt_right">2.320</td> <td class="gt_row gt_right">18.61</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">4</td> <td class="gt_row gt_right">1</td></tr> <tr><td class="gt_row gt_right" style="border-right-width: 3px; border-right-style: solid; border-right-color: lightgrey;">21.4</td> <td class="gt_row gt_right" style="border-right-width: 3px; border-right-style: solid; border-right-color: lightgrey;">6</td> <td class="gt_row gt_right">258</td> <td class="gt_row gt_right">110</td> <td class="gt_row gt_right">3.08</td> <td class="gt_row gt_right">3.215</td> <td class="gt_row gt_right">19.44</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">3</td> <td class="gt_row gt_right">1</td></tr> <tr><td class="gt_row gt_right" style="border-right-width: 3px; border-right-style: solid; border-right-color: lightgrey;">18.7</td> <td class="gt_row gt_right" style="border-right-width: 3px; border-right-style: solid; border-right-color: lightgrey;">8</td> <td class="gt_row gt_right">360</td> <td class="gt_row gt_right">175</td> <td class="gt_row gt_right">3.15</td> <td class="gt_row gt_right">3.440</td> <td class="gt_row gt_right">17.02</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">3</td> <td class="gt_row gt_right">2</td></tr> <tr><td class="gt_row gt_right" style="border-right-width: 3px; border-right-style: solid; border-right-color: lightgrey;">18.1</td> <td class="gt_row gt_right" style="border-right-width: 3px; border-right-style: solid; border-right-color: lightgrey;">6</td> <td class="gt_row gt_right">225</td> <td class="gt_row gt_right">105</td> <td class="gt_row gt_right">2.76</td> <td class="gt_row gt_right">3.460</td> <td class="gt_row gt_right">20.22</td> <td class="gt_row gt_right">1</td> <td class="gt_row gt_right">0</td> <td class="gt_row gt_right">3</td> <td class="gt_row gt_right">1</td></tr> </tbody> </table> </div> --- #### General Functions * [R for Data Science, Programming](https://r4ds.had.co.nz/program-intro.html) * [Software Carpentry - Creating functions](https://swcarpentry.github.io/r-novice-inflammation/02-func-R/) * [Datacarpentry - functions](https://datacarpentry.org/semester-biology/materials/functions-R/) * [UC Business Analytics R Functions guide](https://uc-r.github.io/functions) -- #### Good practices on naming and style * [`tidyverse` style guide on functions](https://style.tidyverse.org/functions.html) -- #### Some `tidyeval` and advanced functions * [`tidyeval` book](https://tidyeval.tidyverse.org/index.html) * [`rlang` 0.4 blogpost](https://www.tidyverse.org/blog/2019/06/rlang-0-4-0/#a-simpler-interpolation-pattern-with) * [Programming w/ `dplyr`](https://dplyr.tidyverse.org/articles/programming.html) * [Using `ggplot2` in packages](https://ggplot2.tidyverse.org/articles/ggplot2-in-packages.html) * [Programming w/ `ggplot2`](https://ggplot2-book.org/programming.html) * [Advanced R, metaprogramming](https://adv-r.hadley.nz/metaprogramming.html) --- ### More realistic internal functions * Connect to and query an internal database w/ `dbplyr` * Take team-specific input data and clean in consistent way * Create a specific plot/table with your colors, logo, and specific data * Generate an entire report * Reusable shiny functions * Scaffolding/wrapper for machine learning tasks -- * ANYTHING you do repeatedly, and want to repeat or test without having to redo manually -- > As Hilary Parker says in her [introduction to packages](https://hilaryparker.com/2014/04/29/writing-an-r-package-from-scratch/): “Seriously, it doesn’t have to be about sharing your code (although that is an added benefit!). It is about saving yourself **time**.” --- ### Internal packages * [Building a team of internal R packages - Emily Riederer](https://emilyriederer.netlify.app/post/team-of-packages/) > More and more organizations are beginning to write their own internal R packages. These internal tools have great potential to improve an organization’s code quality, promote reproducible analysis frameworks, and enhance knowledge management. > > Developers of such tools are often inspired by their favorite open-source tools and, consequently, rely on the design patterns and best practices they have observed. > > Although this is a good starting position, internal packages have unique challenges (such as a smaller developer community) and opportunities (such as an intimate understanding of the problem space and over-arching organizational goals). > > Internal packages can realize their full potential by engineering to embrace these unique features. --- class: center, inverse, middle # Back to package development --- ### Why a package, and not just `source()`? `source()` references a specific `.R` file, reads it all in and executes it (which can include adding a function to the environment). -- `source()`-ing: * Doesn't "know" anything about versioning of the code, ie can be tied to a specific package version * Doesn't have included testing * Doesn't have included documentation * Requires the `.R` file to be copied into every project that needs it * Needs to be changed in _every_ project that needs it * Can be modified or deleted (accidentally) by the end-user --- ### Anatomy of a package * **Metadata** via the `DESCRIPTION`, including the name of the package, description of the package, the version of the package, and any package dependencies -- * **Source code** via `.R` files, that live in the `R/` directory. -- * Special `roxygen` comments inside the `.R` files that describe how the function operates and its arguments, dependencies, and other metadata -- * The **namespace** for the *exported functions* you have written, and the *imported functions* you bring in -- * Tests that confirm your function "works as intended" -- * Other things (installed files, compiled code, data, tutorials, vignettes) --- ### Writing packages, but easier While creating a package can seem intimidating, the `tidyverse` team has spent *years* crafting meta-packages to make life easier to create packages. These are used everyday by 1000s of package developers, and really do make the complicated things easier... with functions! -- * [`devtools`](https://devtools.r-lib.org/): > The aim of devtools is to make package development easier by providing R functions that simplify and expedite common tasks. -- * [`usethis`](https://usethis.r-lib.org/) > usethis is a workflow package: it automates repetitive tasks that arise during project setup and development, both for R packages and non-package projects. --- class: center, middle, inverse # A `demo` package in 5 min --- ### Create a blank package Can call `usethis::create_package()` or use the RStudio GUI. 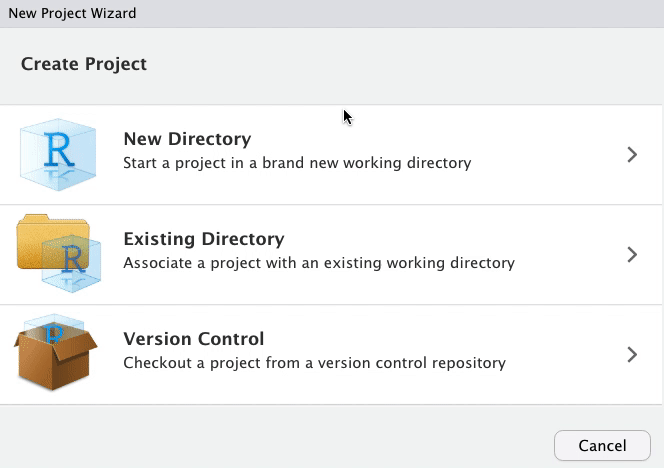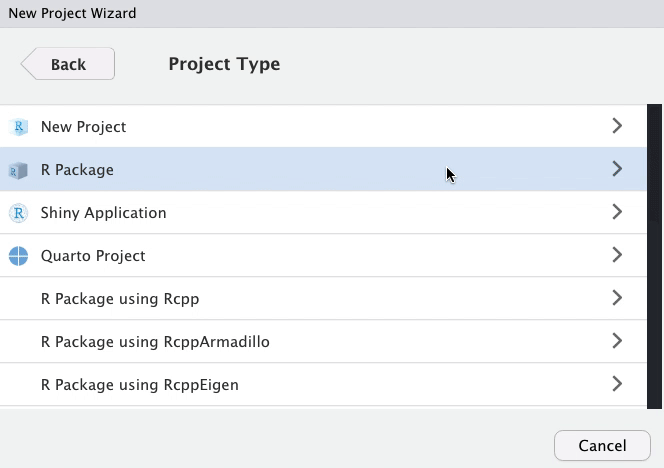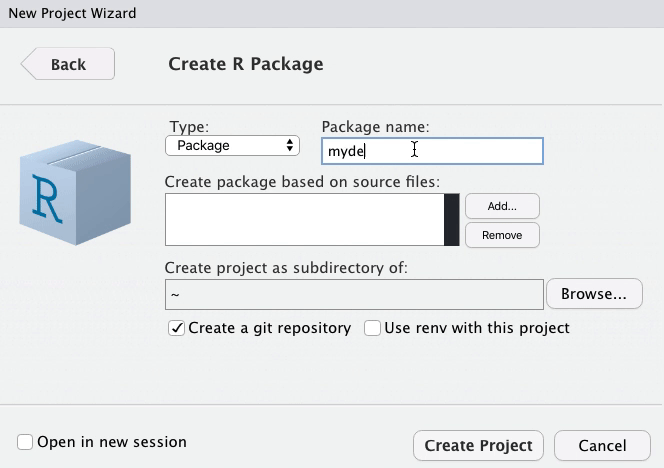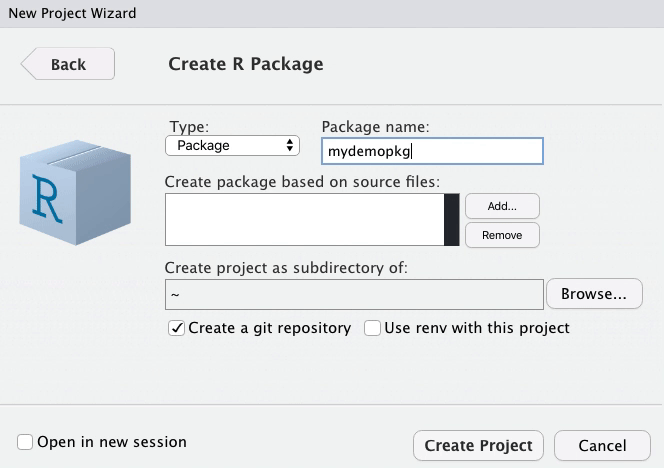 --- class: center, middle, inverse # The "Whole game" --- ### The "Whole game" Load your helper packages and let's go through the process together. ```r # install.packages(c("devtools", "usethis")) library(devtools) library(usethis) ``` --- ### Version Control No shame or shade from us, but you _should_ be using version control for your packages, for your production code, and most, if not all, of your important analysis code. We'll focus on packages for now. This allows your team to collaborate on development, and test changes to the code/packages with code review. -- At RStudio, we typically default to `git` and GitHub, although many folks may use SVN, BitBucket, GitLab, etc. Whatever you're using is great, but note that `usethis` is closely tied to GitHub. -- We can add a `.git` reference, and optionally add GitHub. ```r usethis::use_git() # optionally use github, but you should make a connection # with whatever your version control is # usethis::use_github() ``` -- To read more about Version Control, and specifically how to use `git` with R, please see: [happygitwithr.com](https://happygitwithr.com/) by Jenny Bryan --- ### Start adding our functions This will create a minimal function `.R` file and open it for interactive editing. We can copy our first basic function over and add it to this file. ```r usethis::use_r("square_val.R") ## ✓ Setting active project to '/Users/thomasmock/demopkg' ## • Edit 'R/square_val.R' ## • Call `use_test()` to create a matching test file ``` --- ### Load the function We can load the function with `devtools::load_all()`, this will essentially source the function so we can interactively use it or test it. > `load_all()` has made the function available, although it does not exist in the global workspace. ```r devtools::load_all() ``` -- Once we have confirmed the minimal function is working, we should commit our changes! We can do this via the RStudio `GIT` pane or via the terminal. --- ### Check the function > We have empirical evidence that our function works. But how can we be sure that all the moving parts of the this new package still work? This may seem silly to check, after such a small addition, but it’s good to establish the habit of checking this often. -- We can do this with the `check()` function. -- > `check` automatically builds and checks a source package, using all known best practices ```r devtools::check() ``` -- The output of this function call is quite verbose as it does a LOT of useful checks. The summary at the end tells you how many errors, warnings, or notes are returned. It's important to run this frequently, as you would rather have small moments of friction to fix, rather than doing it much later and having an overwhelming amount of changes to make. --- ### License Now, the function works fine, but we haven't added a license, which `check()` will throw as a warning. -- This may seem silly if you don't intend to publish the package externally to your org, but you should at least define the license so that your wishes are respected as to reuse and ownership of the open source code is clear. -- [R Packages](https://r-pkgs.org/license.html): > Software licensing is a large and complicated field, made particularly complex because it lies at the intersection of programming and law. Fortunately, you don’t need to be an expert to do the right thing: respecting how an author wants their code to be treated as indicated by the license they’ve picked. I default to `use_mit_license()`, but you can read more about licensing: * [Common patterns for licensing R](https://thinkr-open.github.io/licensing-r/rlicense.html) * [R Packages - licensing](https://r-pkgs.org/license.html) * [Choose a License](https://choosealicense.com/) --- ### `document()` your code [R Packages](https://r-pkgs.org/man.html): > Wouldn’t it be nice to get help on our custom function, just like we do with other R functions? This requires that your package have a special R documentation file, man/function_name.Rd, written in an R-specific markup language that is sort of like LaTeX. Luckily we don’t necessarily have to author that directly. -- We can open our existing function, and using RStudio: * `Code > insert roxygen skeleton` I like to do this via the [RStudio Command Palette](https://blog.rstudio.com/2020/10/14/rstudio-v1-4-preview-command-palette/) via `Cmd + Shift + P` There's also a shortcut, `Ctrl + Alt + Shift + R`/`Cmd + Alt + Shift + R` on Windows/Linux or Mac --- ### Let your code breathe on it's own with `roxygen2` * [`roxygen2`](https://roxygen2.r-lib.org/) > The premise of `roxygen2` is simple: describe your functions in comments next to their definitions and `roxygen2` will process your source code and comments to automatically generate `.Rd` files in `man/`, `NAMESPACE`, and, if needed, the Collate field in `DESCRIPTION`. You can read more about `roxygen2` in the helpful [Introduction to `roxygen2`](https://roxygen2.r-lib.org/articles/roxygen2.html). -- `roxygen` items are indicated with special comments (`#'`), ie: ``` #' @param argument A numeric input, that will be squared ``` --- ### Let your code breathe on it's own with `roxygen2` Now, the amount of things you can document can seem overwhelming, but there's only a few things that are specifically necessary. -- * Title of the function w/ **`@title`** * Description of the function purpose with **`@description`** * Documenting the function arguments/parameters with **`@param`** * Specifying it for export with **`@export`** * If it requires external packages, reference the package or specific package functions with **`@import`** and **`@importFrom`** * What does the function return with **`@return`** -- While it's not _required_ it's exceptionally helpful to add a minimal example with **`@examples`** --- ### Basic example ``` #' @title Take a numeric value and square it #' @description This function takes a numeric value, and squares it. #' It is intended to be used as a replacement for `value^2` #' @param x A numeric input, that will be squared #' #' @return a numeric value #' @export #' @examples #' square_val2(4) square_val2 <- function(x){ stopifnot("Input must be numeric" = is.numeric(x)) x^2 } ``` --- ### Now `document()` it Now that we have added the `roxygen2` comments, we can call `devtools::document()` to write out the documentation. -- We can then, just as expected, be able to see the benefit of our labor! ```r ?square_val2 ``` -- While this is great, it's also done some more behind the scenes work. It's added the `square_val2` function as an export in the `NAMESPACE` file. ``` # Generated by roxygen2: do not edit by hand export(square_val2) ``` --- ### Check once, check again Now, we should `devtools::check()` one more time, and make sure we haven't missed anything! -- Also... new `git` commit! Commit early, commit often! --- ### The joy of package installation We can now, within the package project, install! `devtools::install()` when run inside the specific package project, will install the package locally. -- and just as you would expect, we can now load it like any other package! ```r library(demopkg) square_val2(16) #> [1] 256 ``` --- ### Add more functions We can add all of our other functions with a similar workflow... --- ### ~~Add more functions~~ Add our tests We can add all of our other functions with a similar workflow... but for the sake of time we should talk about testing our functions with [`testthat`](https://usethis.r-lib.org/index.html). --- ### Unit testing with `testthat` > Whenever you are tempted to type something into a print statement or a debugger expression, write it as a test instead. > — Martin Fowler -- Up until now, we've only tested our function interactively and checked for package errors via `check()`. -- > We can formalize and expand this with some unit tests via `testthat`. This means we express a few concrete expectations about the correct `square_val2()` result for various inputs. > - [R Packages, testing](https://r-pkgs.org/tests.html) -- [Unit tests are:](https://en.wikipedia.org/wiki/Unit_testing) > typically automated tests written and run by software developers to ensure that a section of an application (known as the "unit") meets its design and behaves as intended...By writing tests first for the smallest testable units, then the compound behaviors between those, one can build up comprehensive tests for complex ~~applications~~ [packages] --- ### Use `testthat` > `usethis` is a workflow package: it automates repetitive tasks that arise during project setup and development, both for R packages and non-package projects. One of the features it provides is the ability to quickly create a `testthat` folder for your package. ```r library(usethis) usethis::use_testthat() ``` --- ### Test structures Tests are organised hierarchically: **expectations** are grouped into **tests** which are organised in files: -- * An **expectation** is the atom of testing. It describes the expected result of a computation: Does it have the right value and right class? Does it produce error messages when it should? An expectation automates visual checking of results in the console. Expectations are functions that start with `expect_`. -- * A **test** groups together multiple expectations to test the output from a simple function, a range of possibilities for a single parameter from a more complicated function, or tightly related functionality from across multiple functions. This is why they are sometimes called unit as they test one **unit** of functionality. A test is created with `test_that()`. -- * A **file** groups together multiple related tests. --- ### `testthat` example Interactively, you can write and check tests. -- ```r devtools::load_all() library(testthat) ``` ```r test_that("Squared values match expectations", { expect_equal(square_val2(2), 2^2) expect_equal(square_val2(4), 4^2) expect_equal(square_val2(16), 16^2) }) ``` ``` ## Test passed 🎊 ``` ```r test_that("Non-numeric or missing inputs should error", { expect_error(square_val2("a"), "Input must be numeric") expect_error(square_val2(factor("a")), "Input must be numeric") expect_error(square_val2(mtcars), "Input must be numeric") expect_error(square_val2(NA), "Input must be numeric") }) ``` ``` ## Test passed 😸 ``` --- ### `testthat` Test structure > A test file lives in `tests/testthat/`. Its name must start with `test`. Here’s an example of a usable test file: -- ```r test_that("Squared values match expectations", { expect_equal(square_val2(2), 2^2) expect_equal(square_val2(4), 4^2) expect_equal(square_val2(16), 16^2) }) #> Test passed 🥇 test_that("Non-numeric or missing inputs should error", { expect_error(square_val2("a")) expect_error(square_val2(factor("a"))) expect_error(square_val2(mtcars)) expect_error(square_val2(NA)) expect_error(square_val2(NULL)) }) #> Test passed 🥳 ``` --- ### What happens if they don't match? -- ```r square_val2("string") ``` ``` ## Error in square_val2("string"): Input must be numeric ``` ```r expect_error(square_val2("string"), "The input must be numeric") ``` ``` ## Error: `square_val2("string")` threw an error with unexpected message. ## Expected match: "The input must be numeric" ## Actual message: "Input must be numeric" ``` -- ```r square_val2(2) ``` ``` ## [1] 4 ``` ```r expect_equal(square_val2(2), 2^3) ``` ``` ## Error: square_val2(2) not equal to 2^3. ## 1/1 mismatches ## [1] 4 - 8 == -4 ``` --- ### More on `testing` While we've covered the basics of unit testing via `testthat`, you'll likely want to read more about it. * [R Packages, Testing chapter](https://r-pkgs.org/tests.html) * [`testhat` documentation](https://testthat.r-lib.org/) * [A long form example of robust testing](https://themockup.blog/posts/2021-03-07-creating-a-custom-gt-function-for-aligning-first-row-text-and-testing-it-with-testthat/) -- Let's move on to more documentation! --- ### Build static `pkgdown` site You can build your very own `pkgdown` site, just like the one for [`devtools`](https://devtools.r-lib.org/reference/build_site.html) by referencing the existing documentation. -- This is super easy, just use `devtools::build_site()` > `devtools::build_site()` is a shortcut for `pkgdown::build_site()`, it generates the static HTML documentation. ```r devtools::build_site() ``` -- This is a nice addendum beyond the built-in documentation within R, and most notably allows you to explore the documentation or share the documentation outside of needing R. As such, you can share information about the package within a team prior to even installing the package. -- The static HTML generated by `pkgdown` can be hosted internally behind authentication via a proper data science tool like RStudio Connect, or publicly on GitHub pages or Netlify. --- ### Hosting `pkgdown` on RStudio Connect * Fully within your firewall * Limited access to your authenticated users * Still _all_ the `pkgdown` goodies! * Can also deploy as static HTML anywhere you want * Example at: [colorado.rstudio.com/rsc/my-pkgdown/](https://colorado.rstudio.com/rsc/my-pkgdown/) ```r rsconnect::deployApp( "docs", # the directory containing the content appFiles = list.files("docs", # the list of files to include as dependencies recursive = TRUE), # (all of the various folders) appPrimaryDoc = "index.html", # the primary file appName = "my-fake-pkgdownsite", # name of the endpoint (unique to your account on Connect) appTitle = "My Pkgdown Site", # display name for the content account = "thomas", # your Connect username server = "colorado.rstudio.com" # the Connect server, see rsconnect::accounts() ) ``` --- ### Reference external packages Now so far, we've not used any additional packages external to base R. To add additional packages, we need to reference them in the `.R` file and in the `DESCRIPTION`. Let's revisit our `gt` example, and show the specific components for change. --- ### A `gt` function ```r gt_add_divider <- function(gt_object, columns, ..., include_labels = TRUE) { stopifnot("Table must be of class 'gt_tbl'" = "gt_tbl" %in% class(gt_object)) gt_object %>% tab_style( # dots include passed named arguments to the internal function style = cell_borders(sides = "right", ...), locations = if (isTRUE(include_labels)) { # columns to affect list(cells_body(columns = {{ columns }}), cells_column_labels(columns = {{ columns }})) } else { cells_body(columns = {{ columns }}) } ) } ``` --- ### A `gt` function, documented We need to specify that we are importing `gt`, so that it's arguments can be referenced in our custom function. We should git commit these changes, and then run `devtools::document()` to rebuild and reference these docs. That will add `gt` as an import in the namespace file, and add `gt_add_divider()` as an export. ```r #' @title Add a column divider to a gt table #' @description Inserts a border to the right of a specific column #' @param gt_object an existing gt_tbl #' @param columns The specific columns where a border will be added #' @param ... Additional arguments passed to `gt::cell_borders()` #' @param include_labels a logical indicating if the border includes column labels #' @importFrom gt %>% #' @import gt #' @export gt_add_divider <- function(gt_object, columns, ..., include_labels = TRUE) {code} ``` --- ### A `gt` function, documented We also need to add `gt` as an import in the `DESCRIPTION` (and git commit), this can be accomplished manually or via `usethis::use_package("gt")`. Note that you can add a specific _version_ of that required package by adding logic after the package name. You have updated this documentation, but you'll need to `devtools::document()` to "bake it in" ``` Package: demopkg Type: Package Title: Our cool new helper function package Version: 0.1.0 Author: Thomas Mock Maintainer: Josiah Parry <jparry@rstudio.com> Description: An amazing package that has lots of cool new features. It also includes a cool new gt function. License: MIT + file LICENSE Encoding: UTF-8 LazyData: true RoxygenNote: 7.1.1 Roxygen: list(markdown = TRUE) Imports: gt (>= 0.3) ``` --- ### Basic workflows within a package REMEMBER! **Smaller** changes committed *frequently* via git. -- * `usethis::use_r()` to add new functions -- * `Ctrl/Cmd + Alt + Shift + R` in RStudio to add `roxygen2` skeleton when inside the function -- * `devtools::load_all()` to interactively test/use your new function -- * `usethis::use_package("gt", type = "Imports")` to add an external package as a dependency -- * `usethis::use_version()` to increment your package version (changes `DESCRIPTION`), note that this commits to git as well! -- * `devtools::document()` to document the package with your various changes -- * `devtools::check()` to check the package -- * If all is well, `devtools::install()` to install it locally --- ### Installation, revisited Now, as far as `devtools::install()`, that takes care of you being able to install it for yourself. -- To share with your colleagues, they can `remotes::install_???` it from your version control, for example I use: > `remotes::install_github("jthomasmock/gtExtras)` -- But you could just as easily use `install_gitlab()` or `install_git()`. -- However, that will only install the most recently released dev version. I could also host the package on an internal CRAN-like repository, or even better, on [RStudio Package Manager](https://www.rstudio.com/products/package-manager/). -- The basic benefit of Package Manager are: * Allowing for multiple versions of the package to be available for installation * Tie to specific package versions and/or specific dates * For all of the packages/dependencies to also be behind your firewall * Install external package dependencies as binary packages (MASSIVELY faster install in Linux) --- ### Wrap-up We've gone through: * The end-to-end process of writing functions with base R and with `tidyeval` * How to create a minimal package * How to add your functions to your package * How to document the package and it's dependencies within R * How to test the functions within your package * How to create public documentation, external to R -- I hope you feel empowered that _you_ can create your own package, and with many best practices. Ultimately, once you get started you'll quickly see that the tooling has been written to make the process very user friendly. --- ### For followup .left-wide[ You can follow the full process in the [R Packages book](https://r-pkgs.org/whole-game.html). It has specific chapters corresponding to the major requirements for building a package. > Welcome to R packages by Hadley Wickham and Jenny Bryan. Packages are the fundamental units of reproducible R code. They include reusable R functions, the documentation that describes how to use them, and sample data. In this book you’ll learn how to turn your code into packages that others can easily download and use. Writing a package can seem overwhelming at first. So start with the basics and improve it over time. ] .right-narrow[ <img src="https://d33wubrfki0l68.cloudfront.net/19c4a5cab01d9bcb1d2edeb63ce5ba0f21870e33/68feb/images/cover.png" title="The cover of the R Packages book" alt="The cover of the R Packages book" width="75%" /> ]